Computational approaches for
discovering hits often involve sorting through many possibilities and examining
a few closely. With luck, some of the predicted molecules will bind to the protein
of interest. However, these don’t always bind for the “right” reason: sometimes
a fragment predicted to bind one way will turn out to bind in quite a different manner. A recent Angew. Chem. Int. Ed.
paper by Gisbert Schneider and colleagues at the ETH in Zürich and SARomics in
Lund reports a possible example.
The researchers were interested
in death-associated protein kinase 3 (DAPK3), which is implicated in several
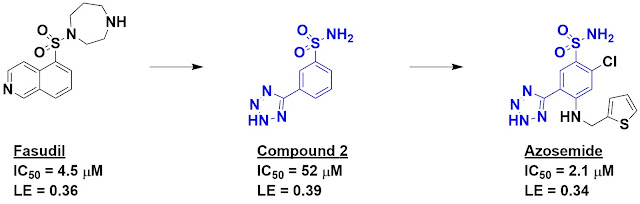
diseases. Previous work had shown that fasudil inhibits this kinase, though it
hits others as well. Fasudil was used as a starting point for de novo fragment
discovery using software called DOGS (Design of Genuine Structures). This is a
scaffold-hopping approach in which virtual chemistry is used to generate
readily accessible alternatives to a starting molecule. In this case, 347 of
the 521 suggested inhibitors were fragment-sized. These were prioritized using
in-house software, and compound 2 – one of the top hits – was chosen for
synthesis and characterization.
Happily, compound 2 turned out to
be fairly potent for its size, with impressive ligand efficiency. It is also
quite different from fasudil (Tanimoto similarity = 0.16). Indeed, while
fasudil is likely to be positively charged at physiological pH, compound 2 is likely to be
negatively charged. Moreover, of 27 other kinases tested, compound 2 hit only
one other with similar potency.
For those who have worked on kinases,
compound 2 does appear unusual. A crystal structure of this molecule bound to
DAPK3 revealed that it sits in the ATP-binding pocket but without making any
conventional hydrogen bond interactions to the so-called hinge region of the
kinase. Although no reported crystal structures show fasudil bound to DAPK3,
structures with other kinases reveal the nitrogen of the isoquinoline moiety
making a hydrogen bond to a backbone amide in this part of the protein.
The software used to prioritize
compound 2 is based not on docking but on machine learning using the ChEMBL
database, and the researchers were interested in what else this fragment might
inhibit. Not surprisingly given the aryl sulfonamide moiety, several carbonic
anhydrases came up, and two were confirmed experimentally.
Interestingly, the diuretic drug
azosemide, whose physiological target is unknown, contains compound 2 as a
substructure, and the researchers found that this molecule inhibits DAPK3 with
low micromolar affinity. It also binds human carbonic anhydrase IX with similar
affinity. The researchers suggest that these targets could at least partially
explain the mechanism of the drug, as well as some of its side effects. It
would be interesting to see cell data against these two targets, as well as the
crystal structure of azosemide bound to DAPK3.



No comments:
Post a Comment